06
Jun
Scientific achievement: Digital PCR became an invaluable tool for GMO quantification
Digital PCR became an invaluable tool for GMO quantification
Products containing genetically modified organisms or GMOs have become more and more complex and diverse since the first genetically modified crop has been released on the market in 1994. Many countries regulate cultivation and trade of GMOs through authorisation systems and mandatory labelling of products containing GMOs above a certain threshold. In Slovenia, as part of the European Union, all products consisting of authorised GMOs need to be labelled when the GMO content exceeds 0.9%. Unauthorised GMOs are forbidden. To enable regulation, a large number of reliable methods for determination of GMO content, or quantification of GMOs in short, have been developed. Still, despite all the efforts, quantification of GMOs in complex samples has proven to be challenging.
Quantification of GMOs is based on counting specific DNA sequences, much like determining the amount of e.g. brown beans in a mixture of brown and grey beans. If we want to calculate the percentage of brown beans, we have to count the number of brown beans as well as brown and grey beans combined. In GMO quantification, we are comparing the number of species-specific sequences, all beans, with sequences specific for a particular GMO, brown beans.
Unlike counting of beans, counting of DNA sequences is not as straightforward. In order to be detected, DNA first needs to be amplified by polymerase chain reaction (PCR) to a sufficient quantity.
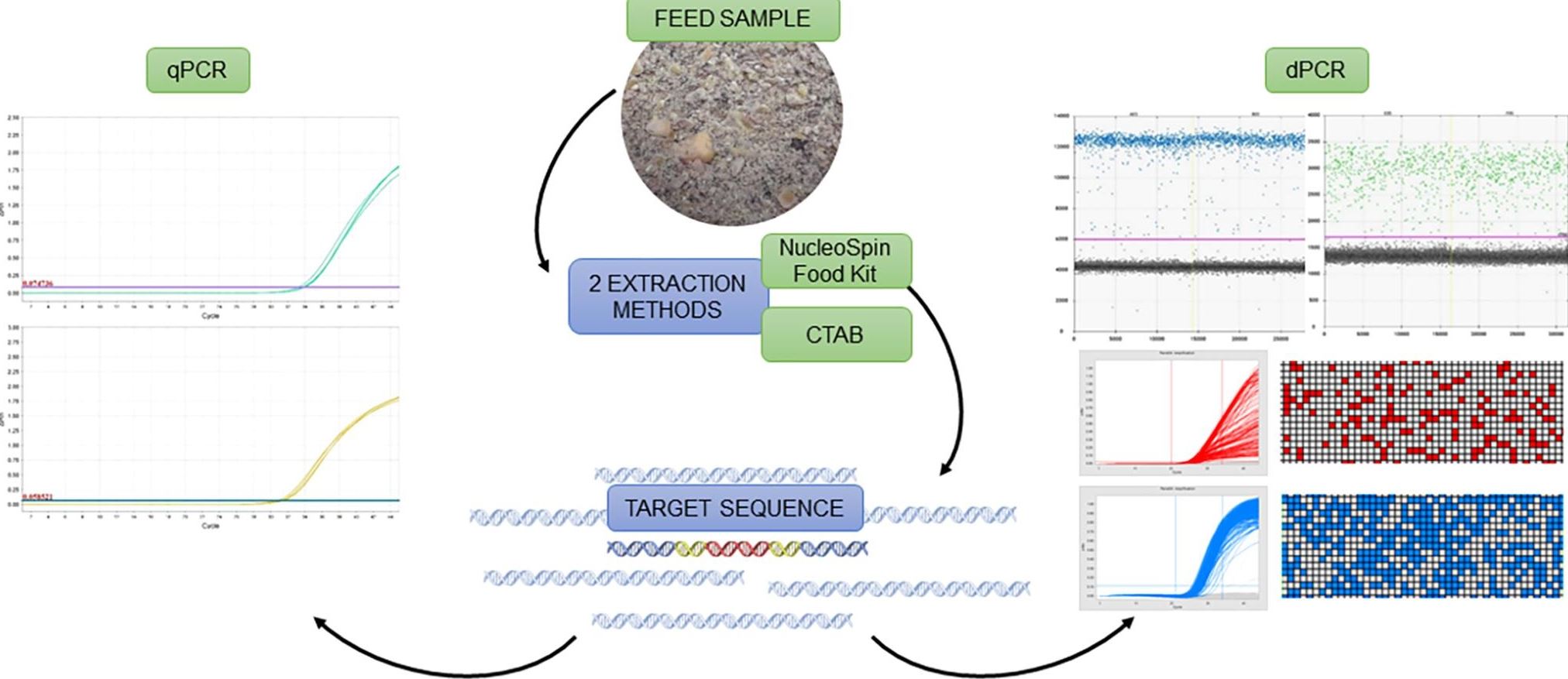
One of the variations of PCR is real-time quantitative PCR or qPCR, an accurate, precise and relatively fast method used in many fields where quantification of DNA or other nucleic acids is needed. Amplification of nucleic acids is achieved with the use of primers (oligonucleotides), which target a specific DNA sequence. From a well-evaluated reference material containing the DNA sequences sought, a calibration curve is prepared. The calibration curve is then compared to the sample curves. Such an approach enables us to determine the amount of target DNA sequences in the sample. In the case of GMO content determination, such analysis is done twice as it is necessary to amplify two different sequences; a sequence characteristic of the GMO and the sequence characteristic of the biological species (species-specific sequence). The proportion of GMOs is then determined by comparing the measured quantities of both sequences.
Unfortunately, qPCR cannot be used for all samples, as it is not reliable enough for samples, which contain a low amount of DNA, or that contain a high concentration of inhibitors that decrease the efficiency of the reaction. In recent years a new approach for quantification of nucleic acids, called digital polymerase chain reaction or dPCR, has been developed. With this approach, a reliable quantification of nucleic acids has significantly increased.
In dPCR, we partition the sample into small sub-samples that contain only one part of the DNA in each sub-sample. In practice, they may be more, but we know that such cases can be properly evaluated using statistical analysis. Then, each of the sub-samples is amplified by PCR. Since we ensure that only the target DNA sequence is amplified, the amplification will only occur in sub-samples containing target DNA.
dPCR does not need a standard curve for DNA quantification. After the reaction, we can count all sub-samples in which the DNA has been amplified and determine the amount of DNA in the sample. For GMO quantification, the amount of DNA characteristic for the GMO and the amount of DNA that is inherent or characteristic for a particular biological species is determined. The ratio of these two numbers corresponds to the proportion of genetically modified DNA in the sample.
Building on a long history of studying methods for nucleic acid quantification, the researchers form the Slovenian National Institute of Biology have proven that dPCR is an invaluable tool in GMO quantification.
In their study, the authors verified the sensitivity and accuracy of the dPCR methods for quantification of Roundup Ready soybean. They showed that different DNA extraction methods have a large effect on qPCR but not on dPCR. This makes dPCR an ideal method for quantification of complex samples that contain more inhibitors of the reaction, contain a low amount of DNA, or where DNA (partially) degraded. In such samples, the content of GMOs cannot be determined by qPCR. However, it can be done with dPCR.
“The technology has proven to be extremely robust, yet sensitive, ideal for quantification of GMOs in complex samples” – said Dr. Alexandra Bogožalec Košir, the first author of the study.
“As we are the National reference laboratory for GMO testing, it was important to find a method that we could use for complex samples, where the GMO content could not be quantified using the standard qPCR approach”– added Dr. Mojca Milavec, the coordinator of the study.
The three dPCR methods introduced in the study have been included in the laboratory’s routine testing scheme.
Reference: A Bogožalec Košir, T Demšar, D Štebih, J Žel, M Milavec (2019) Digital PCR as an effective tool for GMO quantification in complex matrices, Food Chemistry Volume 294, page 73¬¬¬-78.
Contact: Aleksandra Bogožalec Košir, PhD, Research Assistant This email address is being protected from spambots. You need JavaScript enabled to view it. and Katja Ploj, This email address is being protected from spambots. You need JavaScript enabled to view it..