Projects
Biotechnological Hub of the NIB (BTH-NIB)
The purpose of the investment project BTH-NIB is the assurance of the appropriate infrastructural conditions for the use of research and developmental opportunities in the fields of operation of the NIB.
Play Video About project Publication
Biotechnological production of sustainable indole - INDIE
Project coordinator: Dirk Bosch
Coordinator for NIB: Kristina Gruden
Code: C3330-18-252004
Duration: 1.05.2018-30.09.2021
The overall goal of the Indie project is to establish indole production in Corynebacterium glutamicum. This would be an ecological advance as less energy would be used for production, less environmentally harmful waste would be produced and the media would be made from sustainable components. In addition, such production would also be more economical. The bacterium C. glutamicum is safe for the production of substances that are then used as additives in cosmetics or food, as it is one of the organisms recognized as GRAS. There are several approaches to activate indole production in bacteria (Figure 1). Our goal was to find effective triptanase enzymes (TNAs) known to convert tryptophan (Trp) to indole in bacteria. Alternatively, we searched for IGL enzymes responsible for indole production from the Trp precursor, an indop-3-glycerol phosphate (IGP) molecule, in plants. Furthermore, using models of central metabolism, we hypothesize that it is necessary to alter a specific pathway in order for the bacterium to have sufficient precursors for indole synthesis, and accordingly, an additional medium in which to grow the bacteria. Alternatively, Trp could be added directly to the culture medium. In the final phase of the project, industrial production is planned. Optimization of indole extraction from the medium is part of the ongoing project (the end of the project is planned for March 2022).
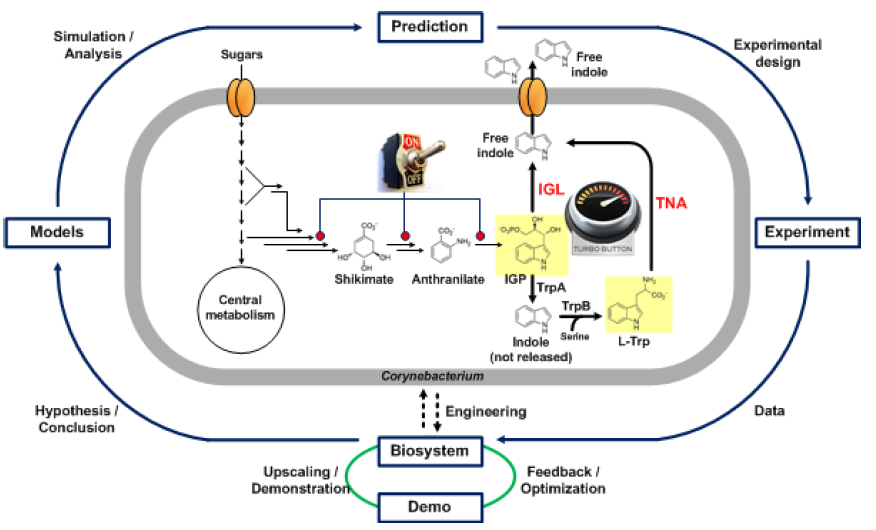
Figure 1: Schematic representation of the activities within the INDIE project.
To search for enzymes of both types, in the first phase of the INDIE project we set up a successful system for rapid assay of the enzyme for indole production. Using the bioinformatics, we found some candidates for both TNA enzymes from bacteria and IGL enzymes from plants. In the last year of the project, we even found that the TrpA protein from C. glutamicum can also take over the function of IGL enzymes when the TrpB enzyme is not present in the bacterium. In parallel, we constructed a complete model of bacterial metabolism and added a regulatory network to it. The predictions obtained using the model guided the modifications of the bacterial genome towards the production of more precursors, the transport of metabolites into the cell, and the transport and synthesis of certain cofactors. Based on these predictions, we also modified the culture medium accordingly. The first generation of bacteria capable of synthesizing indole using the enzyme TNA was grown in 1-liter bioreactors in two ways: with and without the addition of Trp. Indole production was insufficient, so we used a combination of transcriptomics, metabolomics, and modeling to try to identify mechanisms that prevent high production. The results showed transport problems. Indole production was therefore increased by adding an organic solvent to the medium, which redistributed more than 80% of the indole, reducing the saturation of the system with indole and achieving economically acceptable production. The industrial scale production and purification system is still under preparation. We have also established indole production using the enzyme IGL. Through metabolomics, we have found that another by-product is synthesized in addition to indole. In the final phase of the project, we will try to determine its origin using transcriptomics. The cooperation between the project partners has been very successful. We have just started to apply for a project at Horizon Europe HORIZON -CL6-2022-CIRCBIO-02-05 to establish workflows for finding proteins of biotechnological interest in nature, searching for new enzymes in different organisms in the environment.
A short summary of the of NIB's contribution NIB participated in three main activities:
- In work package 2, we used a new bioinformatics workflow to find candidates for direct indole synthesis: By reviewing the literature, we selected plants capable of indole synthesis such that a variant of the IGL enzyme exists in them. Using known IGLs, we identified the motifs required for protein activity and then scanned the entire proteomes of the selected plants for suitable candidates. Of the 10 candidates, 2 proved to be active in bacteria.
- We lead work package 4, which aimed to find mechanisms that block optimal indole production in bacteria. We linked transcriptomic data from indole-producing bacteria using the enzyme TNA with or without the addition of tryptophan to a model of bacterial metabolism and regulation. We used differential analysis of network. We found that transport was problematic, which then led to the addition of organic solvents to the medium and thus the removal of indole from the aqueous phase of the bacterial environment.
- We took care of data management in line with the principles of Open Science and the principles of FAIR. For the needs of the project, we implemented a data management system in synthetic biology. And created a design to record the descriptions of the fermentation experiments. We also developed SeekR scripts to automatically integrate with the FairDomHub API and simultaneously transfer all the project partners' files to this public metadata database.
Web page: https://indie.cebitec.uni-bielefeld.de/
This project is funded by the ERA CoBioTech programme under the European Union’s Horizon 2020 research and innovation programme framework.


Figure 1: Schematic representation of the activities within the INDIE project.
To search for enzymes of both types, in the first phase of the INDIE project we set up a successful system for rapid assay of the enzyme for indole production. Using the bioinformatics, we found some candidates for both TNA enzymes from bacteria and IGL enzymes from plants. In the last year of the project, we even found that the TrpA protein from C. glutamicum can also take over the function of IGL enzymes when the TrpB enzyme is not present in the bacterium. In parallel, we constructed a complete model of bacterial metabolism and added a regulatory network to it. The predictions obtained using the model guided the modifications of the bacterial genome towards the production of more precursors, the transport of metabolites into the cell, and the transport and synthesis of certain cofactors. Based on these predictions, we also modified the culture medium accordingly. The first generation of bacteria capable of synthesizing indole using the enzyme TNA was grown in 1-liter bioreactors in two ways: with and without the addition of Trp. Indole production was insufficient, so we used a combination of transcriptomics, metabolomics, and modeling to try to identify mechanisms that prevent high production. The results showed transport problems. Indole production was therefore increased by adding an organic solvent to the medium, which redistributed more than 80% of the indole, reducing the saturation of the system with indole and achieving economically acceptable production. The industrial scale production and purification system is still under preparation. We have also established indole production using the enzyme IGL. Through metabolomics, we have found that another by-product is synthesized in addition to indole. In the final phase of the project, we will try to determine its origin using transcriptomics. The cooperation between the project partners has been very successful. We have just started to apply for a project at Horizon Europe HORIZON -CL6-2022-CIRCBIO-02-05 to establish workflows for finding proteins of biotechnological interest in nature, searching for new enzymes in different organisms in the environment.
A short summary of the of NIB's contribution NIB participated in three main activities:
- In work package 2, we used a new bioinformatics workflow to find candidates for direct indole synthesis: By reviewing the literature, we selected plants capable of indole synthesis such that a variant of the IGL enzyme exists in them. Using known IGLs, we identified the motifs required for protein activity and then scanned the entire proteomes of the selected plants for suitable candidates. Of the 10 candidates, 2 proved to be active in bacteria.
- We lead work package 4, which aimed to find mechanisms that block optimal indole production in bacteria. We linked transcriptomic data from indole-producing bacteria using the enzyme TNA with or without the addition of tryptophan to a model of bacterial metabolism and regulation. We used differential analysis of network. We found that transport was problematic, which then led to the addition of organic solvents to the medium and thus the removal of indole from the aqueous phase of the bacterial environment.
- We took care of data management in line with the principles of Open Science and the principles of FAIR. For the needs of the project, we implemented a data management system in synthetic biology. And created a design to record the descriptions of the fermentation experiments. We also developed SeekR scripts to automatically integrate with the FairDomHub API and simultaneously transfer all the project partners' files to this public metadata database.
Web page: https://indie.cebitec.uni-bielefeld.de/
This project is funded by the ERA CoBioTech programme under the European Union’s Horizon 2020 research and innovation programme framework.



 Scope of NIB's accreditation is given in the Annex to the accreditation certificate and in the List of accredited methods for detection of GMOs and microorganisms – plant pathogens
Scope of NIB's accreditation is given in the Annex to the accreditation certificate and in the List of accredited methods for detection of GMOs and microorganisms – plant pathogens Holder of National Standard in the Field of Amount of Substance/Bioanalysis of Nucleic Acids/GMOs and Microorganisms
© 2023- National institute of biology, all rights reserved

