NIB
The purpose of the investment project BTH-NIB is the assurance of the appropriate infrastructural conditions for the use of research and developmental opportunities in the fields of operation of the NIB.
Play Video About project Publication
Prof. Dr. Kristina Gruden
Working place: Scientific Counsellor, Head of the Omics Unit
Telephone number: +386 (0)59 23 28 24
Email: kristina.gruden@nib.si
Department: Department of Biotechnology and Systems Biology
Complete bibliography (COBISS)
Bibliography (Google Scholar)The central theme of my research is understanding how plant cells function at the molecular level and based on that, to understand the functioning of whole organism. This requires interdisciplinary research, linking life sciences with computer sciences and mathematics. Our experimental model is the potato (Solanum tuberosum L.) and how it responds to different environmental cues. We focus mainly on potato virus Y (PVY) and the Colorado potato beetle, both of which cause significant economic damage. Lately, we also explore the interaction of potato with its microbiota. We also study response of potato to abiotic stresses such as drought, heat, and flooding, as well as combinations of these. Our main goal is to identify the key components of plant immunity/stress responses that determine whether the outcome of an encounter will be susceptibility, disease, tolerance, or resistance. In our work, we use advanced methods such as analysis of responses over time and space (single-cell and spatial transcriptomics). We combine transcriptomic, metabolomic, hormonomic, proteomic approaches and integrate phenotypic data with various tools, such as integration with knowledge networks (skm.nib.si), which allows us to visualise metabolic and signalling pathways.
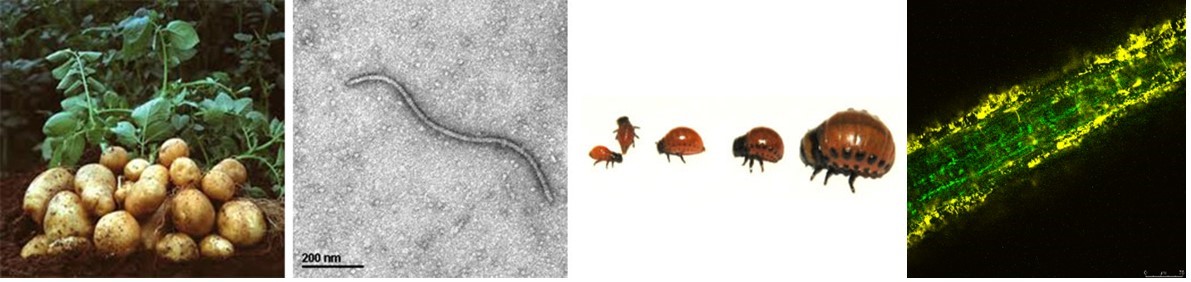
Figure 1: experimental system potato, virus PVY, Colorado potato beetle, epi/endophytes
To facilitate the interpretation of biological data, we have developed several tools:
- SKM: resource integrating knowledge on molecular interactions in plants, specifically protein–protein interactions, protein–promoter interactions, catalysis, transport, post-translational modifications, miRNA–mRNA interactions, and transport
- BoolDog: semiquantitative modelling environment
- DiNAR: allows for visualisation of multiconditional omics data
- Unitato: all annotations of the potato DM genome merged
- QuantGenius: http://quantgenius.nib.si/user/login - a tool for reliable quantification with qPCR, including Fluidigm HT qPCR
- GoMapMan: multispecies visualisation of MapMan gene function ontology
Link SKM >>  |
Link BoolDoG >>  |
Link Unitato >> |
Link DiNAR >>  |
Link GoMapMan >>  |
We have successfully used knowledge networks to integrate multi-omics data, allowing us to monitor events in potato leaves exposed to severe weather conditions: first heat, then a combination of heat and drought, followed by heavy rainfall with short-term flooding.
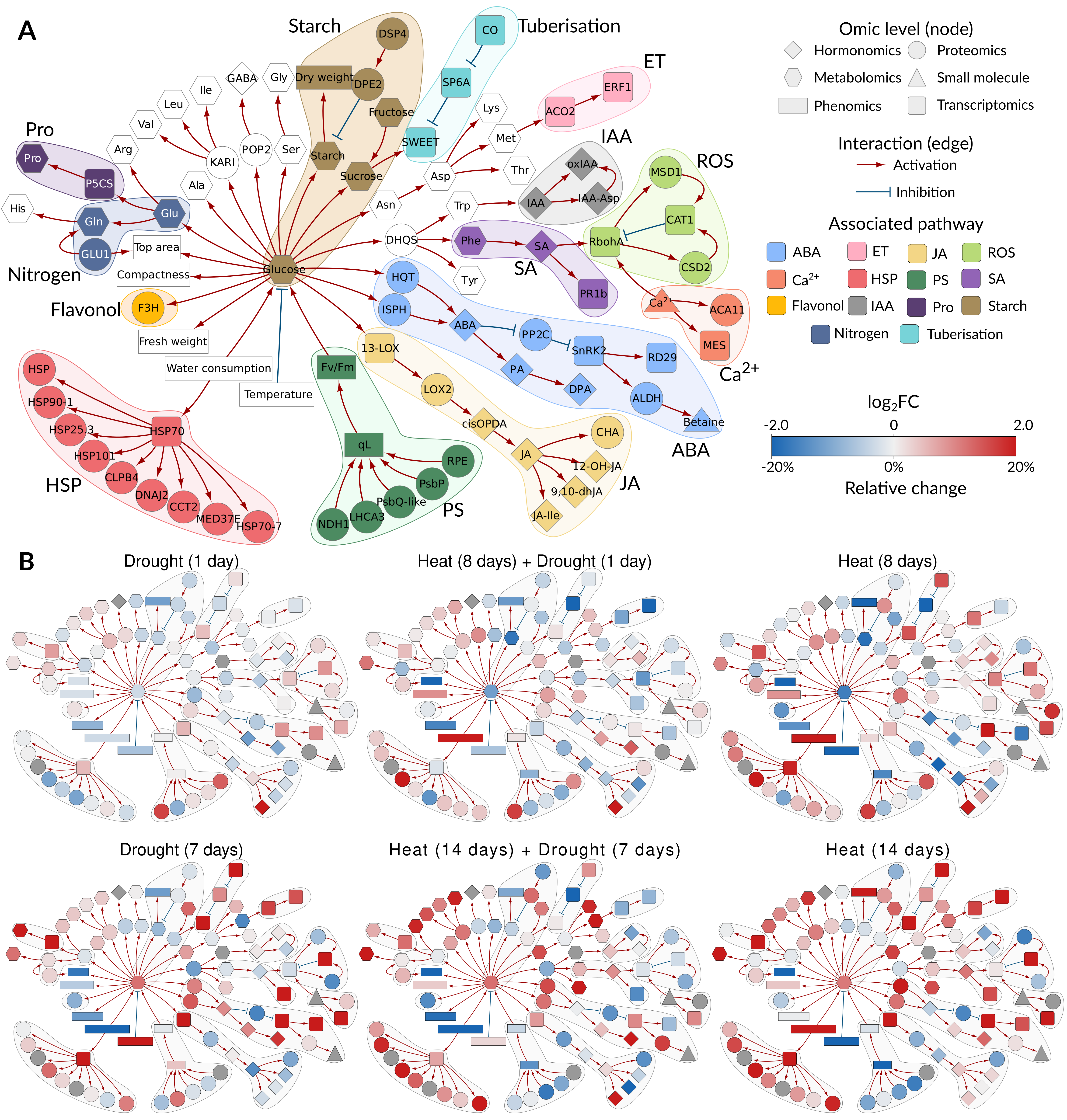
Figure 2: Integration of multi-omics data in a knowledge-based metabolic and signalling network. A) Structure of knowledge network. Individual studied components are coloured according to their function in different pathways. B) To compare the effects of different stresses on the overall state of the plant, we overlaid the knowledge networks with measured changes in component concentration. Nodes are coloured by log2 fold changes (red—increase in stress compared to control, blue—decrease in stress compared with control, grey—measurement not available) shown for 2 time points: sampling day 8 and sampling day 14 for the different stress treatments). Displayed omics measurements were obtained from leaf samples. ABA, abscisic acid; Ca2+, Calcium; ET, ethylene; HSP, heat shock protein; IAA, indole-3-acetic acid (Auxin); JA, jasmonic acid; Pro, proline; PS, photosynthesis; ROS, reactive oxygen species; SA, salicylic acid. From Zagorščak et al.
potatoGEM, is a computational model of the entire metabolic system of the potato. It enables simulations of metabolic changes under different environmental conditions or during attacks by diseases and pests. With this model, it is possible to predict which enzymes and pathways are crucial for resistance or higher yield, thereby guiding the development of new, sustainably cultivated varieties.
Together, the Stress Knowledge Map and potatoGEM form a powerful research platform: the first offers a comprehensive overview of signalling pathways and interactions, while the second enables in-depth simulation of metabolic responses. Combined they allow us to understand plant responses from the moment a threat is perceived to the metabolic reaction, accelerating the development of more resistant plants and sustainable agricultural solutions.
Bleker, C., Ramšak, Ž., Bittner, A., Podpečan, V., Zagorščak, M., Wurzinger, B., Baebler, Š., Petek, M., Križnik, M., van Dieren, A., Gruber, J., Afjehi-Sadat, L., Weckwerth, W., Županič, A., Teige, M., Vothknecht, U. C., & Gruden, K. (2024). Stress knowledge map: a knowledge graph resource for systems biology analysis of plant stress responses. Plant communications, 6(100920), 1–15. https://doi.org/10.1016/j.xplc.2024.100920
Zrimec, J., Correo, S., Zagorščak, M., Petek, M., Bleker, C., Stare, K., Schuy, C., Sonnewald, S., Gruden, K., & Nikoloski, Z. (2025). Evaluating plant growth–defense trade-offs by modeling the interaction between primary and secondary metabolism. Proceedings of the National Academy of Sciences of the United States of America, 122(32, [ ] 2502160122), 1–11. https://doi.org/10.1073/pnas.2502160122
Zagorščak, M., Abdelhakim, L., Rodriguez-Granados, N. Y., Bleker, C., Blejec, A., Zrimec, J., Baebler, Š., Županič, A., Pompe Novak, M., & Gruden, K. (2025). Integration of multi-omics data and deep phenotyping provides insights into responses to single and combined abiotic stress in potato. Plant physiology, 197(4, [ ] 126), 1–42. https://doi.org/10.1093/plphys/kiaf126
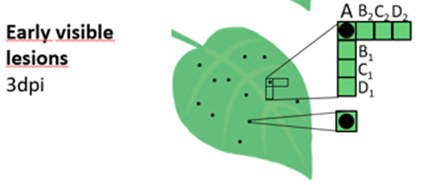
Figure 3: 3D representation of spatial responses to viral infection
Observations of changes in chloroplast redox state revealed that during the hypersensitive response signals spread not only in the cells that die, but also in distant “signalling” cells. These signals are part of the systemic defence network that enables the plant to mount a rapid and coordinated response, with salicylic acid remaining crucial for maintaining the signalling chain.
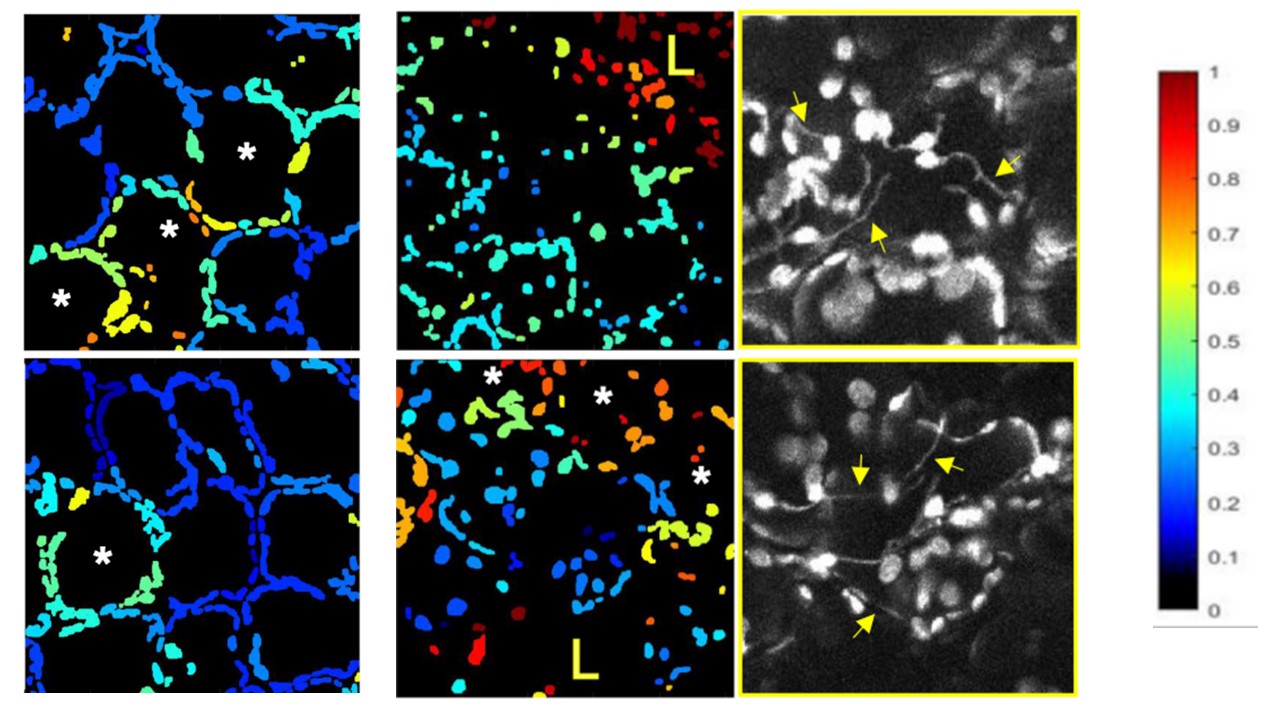
Figure 4: Genetically encoded sensors are fantastic tools for studying responses in plant at cellular and subcellular resolution. Here we used a sensor of chloroplastic redox state pt-roGFP2. We identified signalling cells with oxidised chloroplasts (examples on the left). Interestingly, in the cells surrounding those cells, strong communication between chloroplast and nuclei via stromules is occurring (middle). The legend for relative redox state is shown on the right.
Lukan, T., Županič, A., Mahkovec Povalej, T., Brunkard, J. O., Kmetič, M., Juteršek, M., Baebler, Š., & Gruden, K. (2023). Chloroplast redox state changes mark cell-to-cell signaling in the hypersensitive response. The new phytologist, 237(2), 548–562. https://doi.org/10.1111/nph.18425
Lukan, T., Pompe Novak, M., Baebler, Š., Tušek-Žnidarič, M., Kladnik, A., Križnik, M., Blejec, A., Zagorščak, M., Stare, K., Dušak, B., Coll Rius, A., Pollmann, S., Morgiewicz, K., Hennig, J., & Gruden, K. (2020). Precision transcriptomics of viral foci reveals the spatial regulation of immune-signaling genes and identifies RBOHD as an important player in the incompatible interaction between potato virus Y and potato. The plant journal, 104(3), 645–661. https://doi.org/10.1111/tpj.14953
Some results can also be found via the links below:
Transfer of knowledge into practice
We have developed an innovative biotechnological approach for protecting potatoes against the Colorado potato beetle, using aegerolysins isolated from the edible oyster mushroom. We performed a field trial not using insecticides and the yield of our plants was more than twice as high as that of control plants.
For this invention, we filed a patent application and founded the company Pestevene. The invention was granted the award by the Biotechnical Faculty of the University of Ljubljana in 2024.

Figure 5: Damage to potato plants after Colorado potato beetle attack. Plants protected with proteins from oyster mushrooms (left) and control plants without proteins (right).
Research was published in the eminent journal:
Pogačar, K., Grundner, M., Žigon, P., Coll Rius, A., Panevska, A., Lukan, T., Petek, M., Razinger, J., Gruden, K., & Sepčić, K. (2024). Protein complexes from edible mushrooms as a sustainable potato protection against coleopteran pests. Plant biotechnology journal, 22(9), 2518–2529. https://doi.org/10.1111/pbi.14365


 Scope of NIB's accreditation is given in the Annex to the accreditation certificate and in the List of accredited methods for detection of GMOs and microorganisms – plant pathogens
Scope of NIB's accreditation is given in the Annex to the accreditation certificate and in the List of accredited methods for detection of GMOs and microorganisms – plant pathogens 
