NIB
The purpose of the investment project BTH-NIB is the assurance of the appropriate infrastructural conditions for the use of research and developmental opportunities in the fields of operation of the NIB.
Play Video About project Publication
Prof. Dr. Kristina Gruden
Working place: Scientific Counsellor, Group leader Omics approaches
Telephone number: +386 (0)59 23 28 24
Email: kristina.gruden@nib.si
Department: Department of Biotechnology and Systems Biology
Group leader: Omics approaches
The main topic of my research is focused on understanding of how the cell functions on the molecular level with extrapolation of this knowledge to understanding of organism responses to environmental stress. In parallel I'm also involved in development of methodologies that support such research approach, e.g the systems biology approach. Included are the methods of quantitative molecular biology and also methods that integrate the life sciences with statistic, computer science and mathematics.- Interactions between plants and their pathogens/pests
- Bioinformatics and Systems biology
- Development of quantitative molecular methods – detection of GMOs
See the outcomes of projects ARRS:
- PoDefSig — Setting up the basis for multiscale systems analysis of potato defence signaling. http://projects.nib.si/podefsig/
- Hyp — Spatiotemporal analysis of hypersensitive response to Potato virus Y in potato. http://projects.nib.si/Hyp
Interactions between plants and their pathogens/pests
The interactions between plants and their pests are studied with the final aim of developing new approaches for plant protection. Our pet plant is potato (Solanum tuberosum L.) and we are studying its interaction with most important viral pathogen PVY and insect pest Colorado potato beetle. We are trying to unravel key components determining the outcome of interaction; disease, tolerance or resistance. We are studying the interactions using systems biology approach – data describing the biological system are gathered on multiple molecular levels (gene expression combined with small RNA regulation, metabolomics and proteomics) and merged through data integration tools and modelling. We use time series to get an overview of the process dynamics through quantitative modelling. Further, specific genes are studied using functional genomics tools.

Potato, PVY, Colorado potato beetle, and Functional analysis of genes and their products
IT's all About NETWORKS!
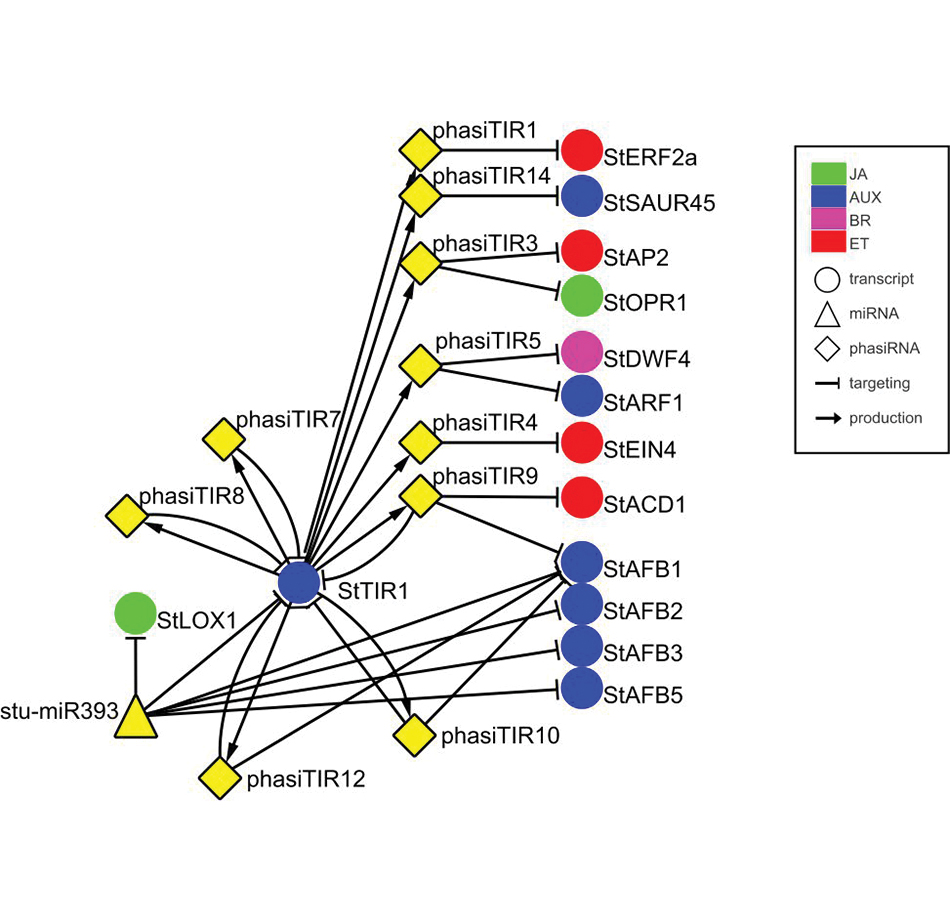
smallRNA regulation of different hormonal modules.
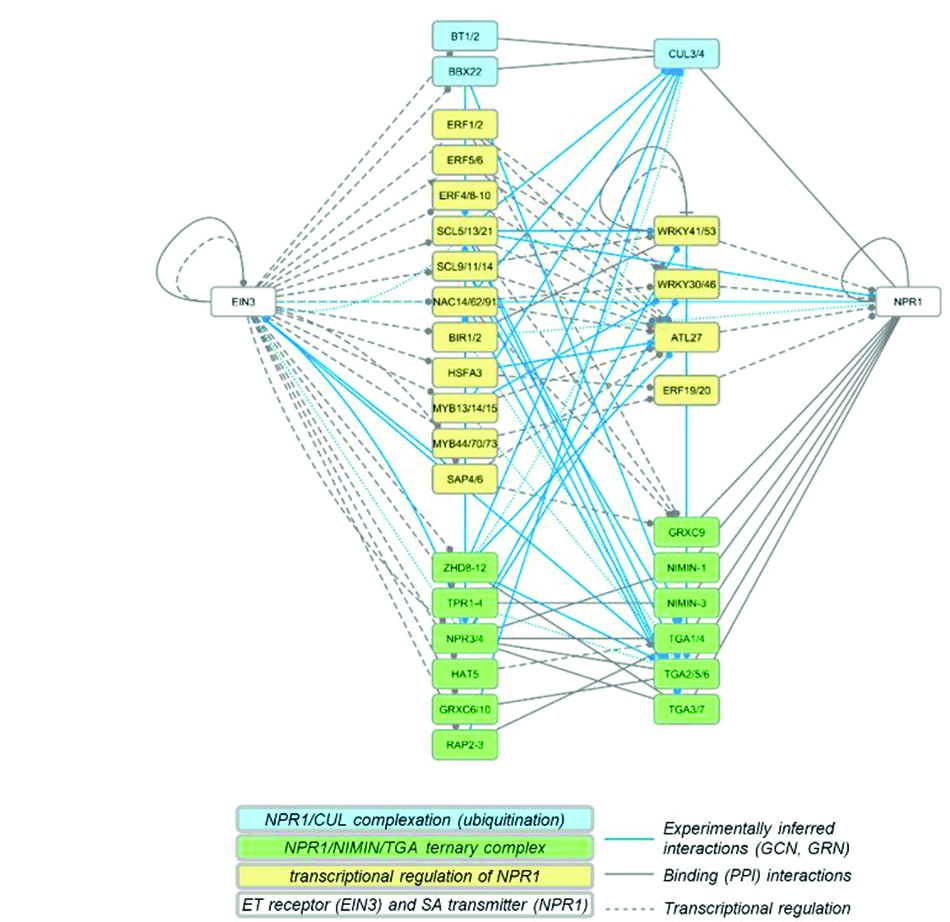
Novel link between ethylene and salicylic acid modules.
Selected publications:
- Ž Ramšak, A Coll, T Stare, O Tzfadia, Š Baebler, Y Van de Peer, K Gruden: Network Modeling Unravels Mechanisms of Crosstalk between Ethylene and Salicylate Signaling in Potato. Plant physiology 178 (1), 488-499 http://www.plantphysiol.org/content/178/1/488
- T Lukan, Š Baebler, M Pompe-Novak, K Guček, M Zagorščak, A Coll, K Gruden. Cell death is not sufficient for the restriction of Potato Virus Y spread in hypersensitive response-conferred resistance in potato. Frontiers in Plant Science 9, 168 https://www.frontiersin.org/articles/10.3389/fpls.2018.00168/full
- SD Schoville, YH Chen, MN Andersson, JB Benoit, A Bhandari, …, M Petek, K Gruden... A model species for agricultural pest genomics: the genome of the Colorado potato beetle, Leptinotarsa decemlineata (Coleoptera: Chrysomelidae) Scientific reports 8 (1), 1931 https://www.nature.com/articles/s41598-018-20154-1
- M Križnik, M Petek, D Dobnik, Ž Ramšak, Š Baebler, S Pollmann, JF Kreuze, J Žel, K Gruden. Salicylic acid perturbs sRNA-gibberellin regulatory network in immune response of potato to Potato virus Y infection. Frontiers in plant science 8, 2192 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5744193/
- T Stare, Ž Ramšak, A Blejec, K Stare, N Turnšek, W Weckwerth, S Wienkoop, D Vodnik, K Gruden. Bimodal dynamics of primary metabolism-related responses in tolerant potato-Potato virus Y interaction. BMC genomics 16 (1), 716 https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-015-1925-2
- Petek M, Rotter A, Kogovšek P, Baebler Š, Mithöfer A and Gruden K. Potato virus Y infection hinders potato defence response and renders plants more vulnerable to Colorado potato beetle attack Molecular Ecology, Volume 23, 5378–5391, 2014 http://onlinelibrary.wiley.com/doi/10.1111/mec.12932/pdf
- Baebler Š, Witek K, Petek M, Stare K, Tušek-Znidaric M, Pompe-Novak M, Renaut J, Szajko K, Strzelczyk-Zyta D, Marczewski W, et al. 2014. Salicylic acid is an indispensable component of the Ny-1 resistance-gene-mediated response against Potato virus Y infection in potato. Journal of experimental botany 65, 1095–1109. http://jxb.oxfordjournals.org/content/early/2014/01/12/jxb.ert447
- Kežar A, Kavčič L, Polák M, Nováček J, Gutiérrez-Aguirre I, Žnidarič MT, Coll A, Stare K, Gruden K, Ravnikar M, Pahovnik D, Žagar E, Merzel F, Anderluh G, Podobnik M. Structural basis for the multitasking nature of the potato virus Y coat protein. Sci Adv. 2019 Jul 17;5(7):eaaw3808. doi: 10.1126/sciadv.aaw3808. https://advances.sciencemag.org/content/5/7/eaaw3808.abstract
Bioinformatics and Systems biology
A structural model of plant defence responses was developed as a first step for systems biology modelling and predictions. The model is based on the knowledge available for the model plant Arabidopsis thaliana and is now being transferred to the crop species. Related to this a database of plant ontologies MapMan was created that allows for easy transfer of knowledge between different plant species besides the more obvious function of a user-friendly and controlled improvement of ontologies and annotations. We additionally created compilation of knowledge dispersed in different public resources from databases to supplements in the literature including PPIs and transcriptional regulation in Arabidopsis and created smallRNA regulatory network for potato.
Interactive model of plant immune signaling that can be opened with Cytoscape, can be found here.
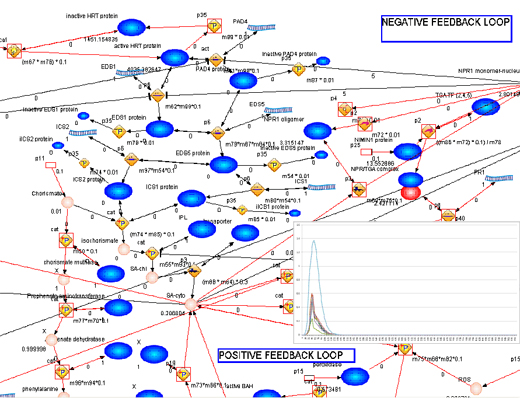
- BioMine: Tool for link discovery in biological databases, currently available for human, mice, rat, arabidopsis, potato, tomato, grapevine
- Database of plant specific gene annotation ontologies, GOMapMan (http://www.gomapman.org/), downloadable updated MapMan ontologies compatible with GSEA and MapMan, available for arabidopsis, rice, potato, tomato, cocoa, sugarbeet and more
- DiNAR: Differential Network Analysis in R, explore the dynamics of response in your multiomics datasets
Selected publications:
M Zagorščak, A Blejec, Ž Ramšak, M Petek, T Stare, K Gruden. DiNAR: revealing hidden patterns of plant signalling dynamics using Differential Network Analysis in R. Plant methods 14 (1), 78 https://plantmethods.biomedcentral.com/articles/10.1186/s13007-018-0345-0
R Schwacke, GY Ponce-Soto, K Krause, AM Bolger, B Arsova, A Hallab, K Gruden, M Stitt, ME Bolger, B Usadel. MapMan4: a refined protein classification and annotation framework applicable to multi-omics data analysis. Molecular plant 2019. https://www.sciencedirect.com/science/article/pii/S1674205219300085
Ramšak Ž, Baebler Š, Rotter A, Korbar M, Mozetič I, Usadel B, Gruden K. 2014. GoMapMan: integration, consolidation and visualization of plant gene annotations within the MapMan ontology. Nucleic acids research 42, D1167–75. doi: 10.1093/nar/gkt1056. http://nar.oxfordjournals.org/content/42/D1/D1167.full
Podpečan V, Ramšak Ž, Gruden K, Toivonen H, Lavrač N. Interactive exploration of heterogeneous biological networks with Biomine Explorer. Bioinformatics. 2019 Jun 24. pii: btz509. doi: 10.1093/bioinformatics/btz509. https://academic.oup.com/bioinformatics/advance-article/doi/10.1093/bioinformatics/btz509/5522368
Development of quantitative methods in molecular biology
We have performed comprehensive research in the field of development of methods for quantitative analysis of nucleic acids. We were testing different alternatives to 5’ nuclease test for following amplification of nucleic acids in real time. The purpose of this research is to get more accurate, more robust or cheaper quantitative methods. Quantitative molecular biological methods, which would be fast and robust as well, are necessary for all types of biological analysis from diagnostics to acquisition of biological data in systems biology concept. Special emphasis was given to decision support system for quality control – QuantGenius. We have applied the knowledge in the area of GMO detection which resulted in identification of potential gaps in detection system due to mutations in inserted DNA sequence.

Selected publications:
Š Baebler, M Svalina, M Petek, K Stare, A Rotter, M Pompe-Novak, K Gruden. quantGenius: implementation of a decision support system for qPCR-based gene quantification. BMC bioinformatics 18 (1), 276 https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-017-1688-7MORISSET, Dany, DEMŠAR, Tina, GRUDEN, Kristina, VOJVODA, Jana, ŠTEBIH, Dejan, ŽEL, Jana. Detection of genetically modified organisms-closing the gaps : to the editor. Nat. biotechnol., 2009, vol. 27, no. 8, 9 str. http://dx.doi.org/10.1038/nbt0809-700, JCR IF (2008): 22.297
MORISSET, Dany, DOBNIK, David, HAMELS, Sandrine, ŽEL, Jana, GRUDEN, Kristina. NAIMA: target amplification strategy allowing quantitative on-chip detection of GMOs. Nucleic acids res., 2008, issue 18, vol. 36, 11 str. http://dx.doi.org/10.1093/nar/gkn524
CANKAR, Katarina, ŠTEBIH, Dejan, DREO, Tanja, ŽEL, Jana, GRUDEN, Kristina. Critical points of DNA quantification by real-time PCR-effects of DNA extraction method and sample matrix on quantification of genetically modified organisms. BMC Biotechnol, 2006, vol. 6, no. 37.


 Scope of NIB's accreditation is given in the Annex to the accreditation certificate and in the List of accredited methods for detection of GMOs and microorganisms – plant pathogens
Scope of NIB's accreditation is given in the Annex to the accreditation certificate and in the List of accredited methods for detection of GMOs and microorganisms – plant pathogens 

