GMOseek: Better control of genetically modified organisms (GMOs) for an improved European food safety
GMOseek is a European project led by NIB and dedicated to the improvement of food safety. The German Federal Office of Consumer Protection and Food Safety (BVL, Germany) and the Food Standard Agency (FSA, UK) fund this project that is part of the ERANET SAFEFOODERA consortium. GMOseek started in June 2009 and is planned to end in June 2011.
GMOseek is a European project led by NIB and dedicated to the improvement of food safety. The German Federal Office of Consumer Protection and Food Safety (BVL, Germany) and the Food Standard Agency (FSA, UK) fund this project that is part of the ERANET SAFEFOODERA consortium. GMOseek started in June 2009 and is planned to end in June 2011.
The GMOseek project is directed towards the challenges of detecting the increasing number of genetically modified organisms (GMOs) approved and unauthorized in the European Union (EU) that that are potentially in the EU food supply. Moreover, GMOseek copes with the time- and cost-efficiency of analytical approaches in order to deliver inexpensive, fast, and broad-based screening tests.
Background
More than 140 different GM plants representing more than 20 plant species have been approved in 55 countries (Figure 1). Not only the presence of GMOs on the market but also their taxonomic (diverse taxon host plants) and biotechnological (diverse genetic constructs) diversity are constantly increasing.
Numerous countries worldwide have adopted legislations dealing with GM crops related issues primarily to ensure food safety. These policies also ensure the consumers' right of choice between GMO and non-GMO derived products. The coexistence of conventional, organic and GM crop production also requires additional traceability. Accurate detection and identification of GMOs are therefore necessary to assure control of GMO traceability. In most GMO detection enforcement laboratories, the standard testing technology is the polymerase chain reaction (PCR, Figure 2) or its quantitative derivative, real-time PCR.

During a typical GMO analysis, a “screening” is performed as a first step in the DNA analysis, in which a minimum set of screening PCR tests (targeting specific genetic elements) should allow to draw conclusions on the absence/presence of as many as possible GM events (Figure 3). Only in case of positive results, a second step using event-specific PCR tests will then specifically identify and/or accurately determine the content of the individual event(s) that is (are) present in the sample. The advantage of such a generic screening based approach is that a minimum set of screening tests is sufficient to cover the maximum number of GM events. In this way, there is no need for performing a high number of event-specific identification tests and thus significant time and costs are saved.
Scientific and technological problem
With the increasing number of EU-approved GM events expected for the next years, the number of screening tests to be carried out will need to be increased accordingly. In addition, laboratories should also be able to distinguish between material derived from authorised and non-authorised GMOs (UGMs), the latest not being assessed for safety in the EU. The increasing number and diversity among GMO traits (Figure 4) raise an urgent challenge: while cost should be reduced, throughput and speed must be increased, in order to keep GMO monitoring time- and cost-affordable for enforcement laboratories.

Research work
The GMOseek project addresses in a very practical and pragmatic way the challenges of GMO screening.
A bioinformatics tool will be developed to automatically extract information from the EUGINIUS project database, an ongoing independent project involving the BVL (Germany) and the RIKILT institute (the Netherlands). From this information, the bioinformatics tool will select the optimal set of new genetic elements that needs to be targeted for detection of a maximum number of GMO events taking in account all currently EU authorized GMOs and (as of yet) UGMs.
As a complement to this computational selection of elements, the second objective of the GMOseek project is the development of new screening real-time PCR methods, targeting new genetic elements (Figure 5). Several genetic elements were already identified for development of screening methods. This part of the GMOseek project will also be fed by the results of the bioinformatics tools that will constantly look at additional genetic elements for which detection method development would be a real added value. Some methods will be developed based on the bioinformatics output.
In addition to developing new screening tools, a significant reduction in number of reactions or tests will be achieved by developing a multiplexed amplification of five genetic target elements in one reaction, by testing, completing and improving the use of existing detection platforms and decision systems.
While PCR is the recommended method for GMO analysis in Europe, disadvantages are its need for expensive equipment and trained personnel and its relatively costly and time-consuming analytical procedure. Therefore, the GMOseek project is also targeting a non-PCR based amplification techniques termed NAIMA and developed by researchers from NIB. The NAIMA method (Morisset et al., 2008, Nucleic Acids Research, 36, e118) provides fast, linear, isothermal amplification of DNA and allows virtually unlimited multiplexing (Figure 6). It is perfectly fit for integrated detection on microarray chip platforms. NAIMA thus significantly increases the speed and throughput while reducing the analysis costs. In this project, the NAIMA method will be developed for the detection of several targets identified as crucial for an efficient GMO screening.

All newly developed methods will be in-house validated and then transferred to a second laboratory as a pre-validation step during the GMOseek project lifetime.
Expected impact
The GMOseek project will provide technical solutions that will be useful to other GMO laboratories and will have an impact in most of the EU member states (and probably even outside Europe). Moreover, the GMOseek project is designed to provide flexible and long lasting tools: the GMOseek algorithm will be built in a way that it adapts to updates of the information contained in the EUGINIUS database.
The GMOseek project will release screening methods allowing cost-efficient and effective detection of all authorized GMOs, and revealing potential presence of UGM. The methods will be ready for full ring-trial validation according to international standards. As an impact, GMOseek should therefore result in a more reliable, standardised and stringent European system of GMO detection.
The detection of authorized GMOs and also UGMs by routine laboratories throughout Europe will be more effective and reliable and if this system can be extended to all enforcement laboratories in the EU, standardisation of GMO screening approach will ensure more harmonised GMO testing of the products released on the European market.
The possibility to better screen for the presence of all possible GMOs will also have an impact on the consumers’ choice between food issued from the biotech industries and conventional/biological food.
NIB researchers involved in the project
Several researchers from the department of Biotechnology and Systems Biology are involved in the GMOseek project. Dr. Dany Morisset is coordinating the GMOseek project and responsible for the NAIMA-microarray based detection development. Pr. Dr. Jana Žel and Dr. Kristina Gruden participate in the bioinformatics tool development, the NAIMA development and the preparation of validation procedures for the methods developed during the project life. MSc. David Dobnik is involved in the NAIMA development.
Other project partners and representatives
Related publications and links
Below are links to publications related to technologies and strategies used in the GMOseek project.
NAIMA and other alternative methods to PCR:
Morisset D, Štebih D, Cankar K, Žel J, Gruden K. Alternative DNA amplification methods to PCR and their application in GMO detection: a review. European Food Research and Technology. 2008, no. 5, vol. 227, pp. 1287-1297. http://dx.doi.org/10.1007/s00217-008-0850-x
Morisset D, Dobnik D, Hamels S, Žel J, Gruden K. NAIMA: target amplification strategy allowing quantitative on-chip detection of GMOs. Nucleic acids res., 2008, issue 18, vol. 36, http://dx.doi.org/10.1093/nar/gkn524
Dobnik D, Morisset D, Gruden K. NAIMA as a solution for future GMO diagnostics challenges. Anal. bioanal. chem., 2009, [online first]. http://dx.doi.org/10.1007/s00216-009-3197-7
Screening real-time PCR methods:
Waiblinger HU, Grohmann L, Mankertz J, Engelbert D, Pietsch K. A practical approach to screen for authorised and unauthorised genetically modified plants. Anal Bioanal Chem. 2009, [online first]. http://dx.doi.org/10.1007/s00216-009-3173-2
Van den Bulcke M, Lievens A, Barbau-Piednoir E, Mbongolombella G, Roosens N, Sneyers M, Casi A.L. A theoretical introduction to "Combinatory SYBR®Green qPCR Screening", a matrix-based approach for the detection of materials derived from genetically modified plants. Anal Bioanal Chem. 2009, [online first]. http://dx.doi.org/10.1007/s00216-009-3286-7
GMOseek is a European project led by NIB and dedicated to the improvement of food safety. The German Federal Office of Consumer Protection and Food Safety (BVL, Germany) and the Food Standard Agency (FSA, UK) fund this project that is part of the ERANET SAFEFOODERA consortium. GMOseek started in June 2009 and is planned to end in June 2011.
The GMOseek project is directed towards the challenges of detecting the increasing number of genetically modified organisms (GMOs) approved and unauthorized in the European Union (EU) that that are potentially in the EU food supply. Moreover, GMOseek copes with the time- and cost-efficiency of analytical approaches in order to deliver inexpensive, fast, and broad-based screening tests.
Background
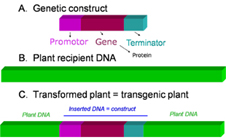
More than 140 different GM plants representing more than 20 plant species have been approved in 55 countries (Figure 1). Not only the presence of GMOs on the market but also their taxonomic (diverse taxon host plants) and biotechnological (diverse genetic constructs) diversity are constantly increasing. 
Figure 1: What is a transgenic plant (GM plant)? A basic genetic construct that contains several genetic elements such as promote, gene and terminator is presented (A). This basic construct is introduced in the plant recipient DNA (B) using diverse techniques. This operation results in a transgenic plant in which the plant endogene DNA is interrupted by the transgene (inserted construct)
Numerous countries worldwide have adopted legislations dealing with GM crops related issues primarily to ensure food safety. These policies also ensure the consumers' right of choice between GMO and non-GMO derived products. The coexistence of conventional, organic and GM crop production also requires additional traceability. Accurate detection and identification of GMOs are therefore necessary to assure control of GMO traceability. In most GMO detection enforcement laboratories, the standard testing technology is the polymerase chain reaction (PCR, Figure 2) or its quantitative derivative, real-time PCR.

Figure 2: Analysis of GMO using conventional PCR. DNA extracted from food sample is amplified using a thermocycler (left); amplified DNA is analysed on poly-acrylamide gel electrophoresis (right)
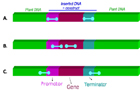
During a typical GMO analysis, a “screening” is performed as a first step in the DNA analysis, in which a minimum set of screening PCR tests (targeting specific genetic elements) should allow to draw conclusions on the absence/presence of as many as possible GM events (Figure 3). Only in case of positive results, a second step using event-specific PCR tests will then specifically identify and/or accurately determine the content of the individual event(s) that is (are) present in the sample. The advantage of such a generic screening based approach is that a minimum set of screening tests is sufficient to cover the maximum number of GM events. In this way, there is no need for performing a high number of event-specific identification tests and thus significant time and costs are saved.

Figure 3: GMO PCR analysis steps. Generally, a first screening step using PCR screening tests is applied. Screening step usually targets genetic elements that are common to several GMOs such as regulatory elements (promoters, terminators…) (A), genes or typical junctions between two genetic elements (B). Depending on the outcome of the screening step, identification step is performed with the so-called event-specific PCR tests that target the unique junction between the plant DNA and the transgene (C)
Scientific and technological problem
With the increasing number of EU-approved GM events expected for the next years, the number of screening tests to be carried out will need to be increased accordingly. In addition, laboratories should also be able to distinguish between material derived from authorised and non-authorised GMOs (UGMs), the latest not being assessed for safety in the EU. The increasing number and diversity among GMO traits (Figure 4) raise an urgent challenge: while cost should be reduced, throughput and speed must be increased, in order to keep GMO monitoring time- and cost-affordable for enforcement laboratories.
Figure 4: Projected number of events in GM crops worldwide by crop (B) and by trait (A). Source: Alexander J. Stein and Emilio Rodríguez-Cerezo, 2009, The global pipeline of new GM crops: implications of asynchronous approval for international trade, EUR 23486 EN – Joint Research Centre – Institute for Prospective Technological Studies, Luxembourg: Office for Official Publications of the European Communities
Research work
The GMOseek project addresses in a very practical and pragmatic way the challenges of GMO screening. A bioinformatics tool will be developed to automatically extract information from the EUGINIUS project database, an ongoing independent project involving the BVL (Germany) and the RIKILT institute (the Netherlands). From this information, the bioinformatics tool will select the optimal set of new genetic elements that needs to be targeted for detection of a maximum number of GMO events taking in account all currently EU authorized GMOs and (as of yet) UGMs.
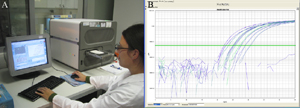
As a complement to this computational selection of elements, the second objective of the GMOseek project is the development of new screening real-time PCR methods, targeting new genetic elements (Figure 5). Several genetic elements were already identified for development of screening methods. This part of the GMOseek project will also be fed by the results of the bioinformatics tools that will constantly look at additional genetic elements for which detection method development would be a real added value. Some methods will be developed based on the bioinformatics output.

Figure 5: Analysis of GMO using real-time PCR. DNA extracted from food sample is amplified using a thermocycler allowing real-time monitoring of the reaction (A); amplified DNA is directly analysed “on screen” using dedicated software (B)
In addition to developing new screening tools, a significant reduction in number of reactions or tests will be achieved by developing a multiplexed amplification of five genetic target elements in one reaction, by testing, completing and improving the use of existing detection platforms and decision systems.
While PCR is the recommended method for GMO analysis in Europe, disadvantages are its need for expensive equipment and trained personnel and its relatively costly and time-consuming analytical procedure. Therefore, the GMOseek project is also targeting a non-PCR based amplification techniques termed NAIMA and developed by researchers from NIB. The NAIMA method (Morisset et al., 2008, Nucleic Acids Research, 36, e118) provides fast, linear, isothermal amplification of DNA and allows virtually unlimited multiplexing (Figure 6). It is perfectly fit for integrated detection on microarray chip platforms. NAIMA thus significantly increases the speed and throughput while reducing the analysis costs. In this project, the NAIMA method will be developed for the detection of several targets identified as crucial for an efficient GMO screening.

Figure 6: NAIMA method. NAIMA multiplex amplification results in production of multiplex RNA molecules (A) that are then analysed after hybridization on microarray (B)
All newly developed methods will be in-house validated and then transferred to a second laboratory as a pre-validation step during the GMOseek project lifetime.
Expected impact
The GMOseek project will provide technical solutions that will be useful to other GMO laboratories and will have an impact in most of the EU member states (and probably even outside Europe). Moreover, the GMOseek project is designed to provide flexible and long lasting tools: the GMOseek algorithm will be built in a way that it adapts to updates of the information contained in the EUGINIUS database. The GMOseek project will release screening methods allowing cost-efficient and effective detection of all authorized GMOs, and revealing potential presence of UGM. The methods will be ready for full ring-trial validation according to international standards. As an impact, GMOseek should therefore result in a more reliable, standardised and stringent European system of GMO detection.
The detection of authorized GMOs and also UGMs by routine laboratories throughout Europe will be more effective and reliable and if this system can be extended to all enforcement laboratories in the EU, standardisation of GMO screening approach will ensure more harmonised GMO testing of the products released on the European market.
The possibility to better screen for the presence of all possible GMOs will also have an impact on the consumers’ choice between food issued from the biotech industries and conventional/biological food.
NIB researchers involved in the project
Several researchers from the department of Biotechnology and Systems Biology are involved in the GMOseek project. Dr. Dany Morisset is coordinating the GMOseek project and responsible for the NAIMA-microarray based detection development. Pr. Dr. Jana Žel and Dr. Kristina Gruden participate in the bioinformatics tool development, the NAIMA development and the preparation of validation procedures for the methods developed during the project life. MSc. David Dobnik is involved in the NAIMA development.Other project partners and representatives
- Centre Wallon de Recherches Agronomiques (CRA-W, Belgium), http://www.cra.wallonie.be/, Gilbert Berben (This email address is being protected from spambots. You need JavaScript enabled to view it.)
- Scientific Institute of Public Health (IPH, Belgium), http://www.iph.fgov.be/, Nancy Roosens (This email address is being protected from spambots. You need JavaScript enabled to view it.)
- Own Equity of the Institute for Agricultural and Fisheries Research (EV ILVO, Belgium), http://www.ilvo.vlaanderen.be/, Isabel Taverniers (This email address is being protected from spambots. You need JavaScript enabled to view it.)
- Bavarian Health and Food Safety Authority (LGL, Germany), http://www.lgl.bayern.de/, Ulrich Busch (This email address is being protected from spambots. You need JavaScript enabled to view it.)
- EU Joint Research Centre - Institute of Health and Consumer Protection (JRC-IHCP, EC), http://mbg.jrc.ec.europa.eu/, Marc van Den Bulcke (This email address is being protected from spambots. You need JavaScript enabled to view it.)
Related publications and links
Below are links to publications related to technologies and strategies used in the GMOseek project.
NAIMA and other alternative methods to PCR:
Morisset D, Štebih D, Cankar K, Žel J, Gruden K. Alternative DNA amplification methods to PCR and their application in GMO detection: a review. European Food Research and Technology. 2008, no. 5, vol. 227, pp. 1287-1297. http://dx.doi.org/10.1007/s00217-008-0850-x
Morisset D, Dobnik D, Hamels S, Žel J, Gruden K. NAIMA: target amplification strategy allowing quantitative on-chip detection of GMOs. Nucleic acids res., 2008, issue 18, vol. 36, http://dx.doi.org/10.1093/nar/gkn524
Dobnik D, Morisset D, Gruden K. NAIMA as a solution for future GMO diagnostics challenges. Anal. bioanal. chem., 2009, [online first]. http://dx.doi.org/10.1007/s00216-009-3197-7
Screening real-time PCR methods:
Waiblinger HU, Grohmann L, Mankertz J, Engelbert D, Pietsch K. A practical approach to screen for authorised and unauthorised genetically modified plants. Anal Bioanal Chem. 2009, [online first]. http://dx.doi.org/10.1007/s00216-009-3173-2
Van den Bulcke M, Lievens A, Barbau-Piednoir E, Mbongolombella G, Roosens N, Sneyers M, Casi A.L. A theoretical introduction to "Combinatory SYBR®Green qPCR Screening", a matrix-based approach for the detection of materials derived from genetically modified plants. Anal Bioanal Chem. 2009, [online first]. http://dx.doi.org/10.1007/s00216-009-3286-7