Systems Biology
Interactions of plants with multiple stressors
Tripartite interaction: potato – potato virus Y (PVY) – potato Colorado beetle (CPB)
When insects attack, plants start to produce numerous antinutritional defence substances, such as enzyme inhibitors and polyphenol oxidases. These can cause growth retardation and increased mortality of insects. After ingestion of antinutritional compounds, insects respond by up-regulating the expression of existing enzymes and in some cases by expressing inhibitor-insensitive digestive enzymes. Our aim is to explore and understand molecular interactions between potato and CPB (CPB, Fig. 3) and to indentify key genes and proteins that enable Colorado potato beetle to counteract plant's defences and normally develop on potato plants.
Because crops in the field can be simultaneously attacked by herbivores as well as pathogens, we have started to investigate a tripartite interaction between potato, Colorado potato beetle and Potato virus Y (PVY). We used RNAseq to study gene expression in the potato leaves and the gut of the CPB larvae, and CG-MS-based metabolomics to study the release of potato volatile compounds. We have shown that susceptible potato plants activate the responses to PVY infection that counteracts their antiherbivore defences. We have shown that after CPB attack ethylene and auxin signalling pathways are attenuated in PVY-infected plants causing lower accumulation of antinutritional compounds. PVY infection therefore renders plants more vulnerable to subsequent CPB attack. Additionally, Colorado potato beetle and PVY both modulate the release of potato volatiles, which can cause priming of neighbouring potato plants or affect the behaviour and response of other associated organisms in the environment. In this way, combinations of different biotic stressors can have broader agro-ecological consequences.

Figure 3: Colorado potato beetle (CPB)
Development of Systems biology methodologies
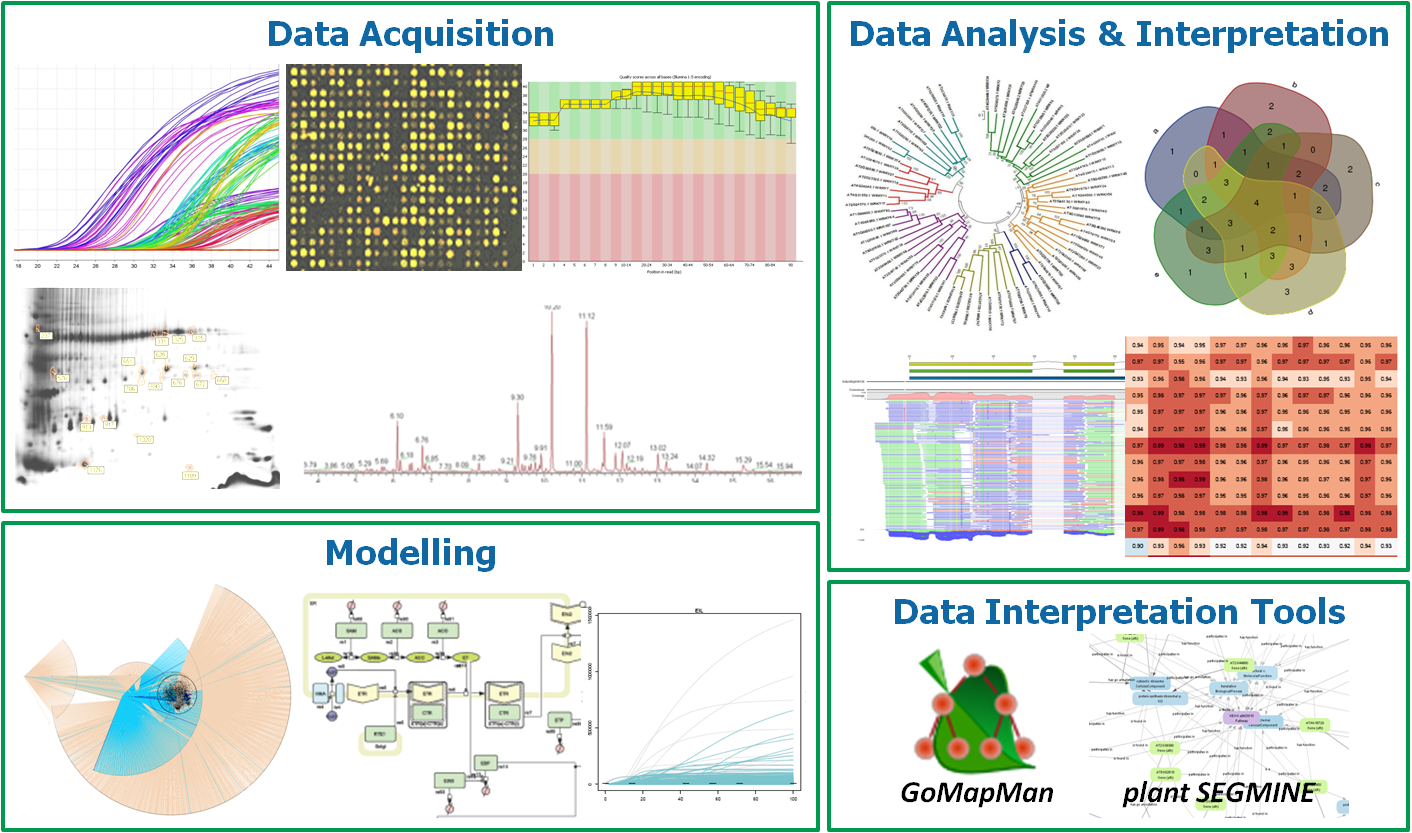
Figure 4: A schematical representation of the approaches taken within the group.
Data Acquisition
Systems biology treats biological problems as a whole. Here, transcriptomics with methods of next generation sequencing, DNA microarrays and quantitative real-time PCR is successfully included. In the group, we use these methods for studying processes in various biological topics. We complement the results acquired on the transcriptomics level with the proteomics level (2D-PAGE in connection with mass spectrometry, shot-gun MS and MRM and analysis of enzyme activities (tight collaboration with Prof. Weckwert, University of Vienna). In co-operation with the Max-Planck Institute for Chemical Ecology (Dr. Axel Mithoefer, Jena, Germany), we are also introducing the analysis of volatile compounds, which plants release in communication with the environment. Within the biochemical and cytological research, we study the involvement of plant growth regulators (cytokinins, jasmonic and salicylic acid), oxygen radicals, photosynthetic pigments and some proteins in the defence response of plants.
Data Analysis & Interpretation
All methodologies representing elements of the systems approach are tightly connected with the acquisition of a large quantity of data, which represents a challenge not only for the biologists but also for statisticians and bioinformatitians. Therefore, careful experimental planning, taking into account different perspectives, is crucial for the appropriate final result.
At the Department, we build statistical models (regression and linear models) and apply multivariate analysis approaches (principal component analysis and discriminant analysis) to the posed biological questions. In addition to statistical methods, we also apply data mining methods, decision trees among others. We additionally develop new algorithms and workflows appropriate for the particular biological questions and hypotheses, if needed. We have especially directed ourselves towards the analysis of transcriptomic data, which includes data quality control, data pre-processing, statistical data analysis and result visualisation.
Data Interpretation Tool Development
To integrate the data into its biological context, the group is also involved in tasks of merging prior knowledge with experimental data. This is achieved through development of tools that allow for high-throughput meta-analyses or or preparing our data for other publically available analysis software. Based on the MapMan ontology the GoMapMan application (www.gomapman.org) has been developed, combining ontological functional descriptors with orthologue information to allow easier transfer between a model organism (Arabidopsis) and various crop plants. Apart from organising genes and their ontology assignments, GoMapMan also offers input files for other software, such as Gene Set Enrichment Analysis (GSEA) or MapMan tool. In other related projects, plantSEGMINE, a tool to assist in creating and sharing data analysis workflows has been developed in cooperation with the Jozef Stefan Institute.
Modelling
A structural model of plant immune signalling (PIS) was already developed as a first step for systems biology modelling and predictions. It includes the signalling components of three important hormones of plant defence, jasmonic acid, salicylc acid and ethylene. Interactive model that can be opened with Cytoscape, can be found here. We are currently fine-tuning the model with the knowledge available in public domain, e.g. protein-protein interactions, transcriptional regulatory network and miRNA regulatory network. The final goal of the network construction is to allow for dynamical modelling and new hypothesis generation to satisfy the original systems biology paradigm.
Abscission in tomato
Abscission is a highly temporally and spatially regulated process in which various organs, including leaves, flowers and flower organs or fruits, are separated from the mother plant. The basis for organ abscission is considered to be a cell-separation process, which occurs specifically in the pre-formed abscission zone (AZ) tissue located at the base of the organ to be shed. The AZ is morphologically and physiologically distinct from neighbouring cells and includes smaller-sized, cytoplasmically dense cells forming from 1~50 cell layers in different plants. We have already demonstrated that the abscission-related processes in tomato leaf abscission zone occur asymmetrically between the AZ proximal and distal sides. Our findings identify program cell death (PCD) as a key mechanism that occurs asymmetrically during normal progression of abscission, and suggest an important role for LX in this PCD process. In the current study we investigate several enzymes with the presumed role in leaf and flower tomato abscission on the transcriptome, structural and ultarstructural levels.